User-guide last updated: 11 June, 2024
User-guide first published: 2 February, 2024
About the ARGA index
The Australian Reference Genome Atlas (ARGA) indexes a range of genomic and genetic datasets generated from biological species relevant to Australia. The index includes publicly available data from several genetic data repositories and focuses on animals, plants, fungi, algae and protozoans; currently, viruses and bacteria are not included in the ARGA index. New data sources are added regularly, and already included data sources are refreshed via a scheduler. A full list of data sources, with hyperlinks to their original online location, is provided on the Data Sources page; this page also includes metadata about the last date of indexation and number of records included. The Data Sources page can be accessed from the global menu bar at the top of the page in the ARGA app.
ARGA has a defined scope which covers only eukaryote species, and includes those species known to occur in Australia and external territories, as well as additional species which may be of relevance to Australia’s interests in biosecurity, agriculture, commerce and trade, and the wider evolutionary origins for Australia’s unique biodiversity. The taxonomic backbone that supports the ARGA search engine is built using a mixture of primary sources to create a single unified names index. The ARGA names index relies on authoritative sources to provide currently accepted names for species, and is drawn chiefly from the Australian National Species Lists, which include the Australian Plant Census and the Australian Faunal Directory. Additional sources have been incorporated for species not contained within these lists. A full list of sources can be accessed on the Data Sources page. Sources are indicated for each taxonomic name on its relevant page within the ARGA app.
Important note about data usage and limitations
Attributions of data to individual taxonomic species as displayed in the ARGA index are inherited from source datasets. They are not cross-checked by ARGA for the underlying scientific validity of the taxonomic identification, and as such, users of ARGA should be aware that the aggregation of a specific datum within a species page depends on the identification of that material in the source data (i.e. if a sequence is ascribed to an “incorrect” (as in misidentified) taxonomic name in the source data, it will remain in the “wrong” species). Similarly, data for a particular species may not appear in a search if it has been deposited under a name which is no longer in scientific usage. ARGA has built into its name index a list of known synonyms from reputable sources (and as reflected on the National Species Lists and the Atlas of Living Australia), and while every effort has been taken to ensure currency of this list, it should be acknowledged that these lists are built from taxonomic opinion and precedent, and may therefore represent a given taxon under an unexpected name, and also that no list can be exhaustive. Users are encouraged to explore the nomenclatures used by the National Species Lists and Atlas of Living Australia, as well as those provided at the Global Biodiversity Information Facility (GBIF) and Catalogue of Life. If you believe that a name is inaccurate, please reach out to these data providers directly (with peer-reviewed documentation), or you may also contact us, and we will work with you to access the data providers so that they may assist with having any updated names reflected in the various lists and catalogues of species.
ARGA makes no claims about the quality of any of the genomic data that it indexes. ARGA provides users with metadata to assist them to assess the usefulness of any given data for their own specific context. Users are encouraged to explore all of the data for any given taxon and view the metadata to help them in their own assessments of the applicability of a datum for inclusion and reuse in any downstream studies. Attributions for source data are provided with each piece of data included in the index. Users should respect the data licensing agreements as indicated for each datum, and cite all used data accordingly in any derivative works. Users are free to share (under a CC-BY 4.0 licence) any of the aggregated metrics presented by ARGA on the higher taxon summary pages, and are requested to cite ARGA as a source for these metrics. Please refer to the Data sources and citations section below for instructions on citing ARGA and source materials.
Searching in ARGA
Searches can be made from the search bar on the homepage. Search terms can relate to taxonomic, genomic, or specimen data attributes. You can combine multiple search terms to craft more specific searches. See query syntax for more information on how to build a search around defined parameters.
Searching by scientific or common names for species and higher groups
ARGA supports searching for species using both common names and scientific names, and at any taxonomic level. The simplest way to commence a search is to type in the name of your species of interest, for example, you could type {koala} to obtain information about the genomic data discoverable for koalas within the ARGA index. If you know the Latin name of your species, you could achieve the same search results by typing {Phascolarctos cinereus} into the search bar and pressing the {Enter} or {Return} key.
Tip: Make sure any scientific names follow the correct capitalisation, e.g. {Aves} or {Thalassarche bulleri}.
The ARGA names index enables searching for animals and protists using any of the following ranks: kingdom, phylum, class, order, family and genus. For plants, algae and fungi, searching has been enabled for the ranks: regnum, division, classic, ordo, familia and genus. A search using a higher taxon will yield results for all of the indexed members of that group, so a search for {Acacia} will return all of the species reports, genomic data and specimen records that include the term {Acacia} somewhere in the data record. Searching using a species-rich group name, like {Acacia} or {Lutjanus} may yield a large number of results. If you have searched at a high taxonomic level (like phylum or division), you may like to deploy additional filters or search terms to limit your query. Alternatively, you may like to build more complex queries, like {genus:Acacia} or {genus:Lutjanus}, to restrict the returned list of the set of all of the species reports within the ARGA index (i.e. only the aggegrated species reports will be returned, not individual data items) (see query syntax below for additional constructions, strategies and methods for search limitation).
Hint: If you are searching with a species name (or other scientific name) that doesn’t yield any results, check whether it has been described under a different name in an authoritative taxonomic resource.
If you do not know the scientific name of your target species, you can still search using a common name, like {frog} or {kangaroo}, but please be aware that these results will be less specific. For instance, the search {kangaroo} will bring back results for any species with {kangaroo} in the data, such as Amblyomma triguttatum Koch, 1844 (the kangaroo hard-tick) or Requena kangaroo Rentz, 2001 (a grasshopper from Kangaroo Island).
Searching by genomic data identifier
Accession numbers (or equivalent repository-specific sequence identifiers) can be used to search for specific sequences. Repository names, such as {NCBI GenBank} or {BOLD}, can be used as search terms to return data exclusively from specified sources. You can also specify the type of data you are searching for in the search bar to return only genome assemblies {data_type:Genome}, or single loci {data_type:Locus}.
Hint: Use the versioning decimal suffix when searching for INSDC accessions, e.g. {GCA_XXXXXXXXX.2}.
Searching with specimen data attributes
ARGA provides specimen records as a separate data type, and these can be queried using relevant data attributes, including the institution/institution code and collection in which the specimen was deposited and the recorded collector and identifier. You can search for specimens explicitly {data_type:Specimen}, and for specimens indexed from a specified repository.
Tip: ARGA indexes Australian museum specimen records provided to the Atlas of Living Australia by the Online Zoological Collections of Australian Museums (OZCAM). To search for museum-accessioned specimens, use {data_source:OZCAM}.
Query syntax
ARGA’s search accepts specifications on a range of data attribute fields. These can be defined with the colon operator, according to the syntax {attribute}:{value}. Table I below lists the full list of fields which can be used to formulate queries.
| Data attribute field name | Description | Example |
|---|---|---|
| Taxonomic attributes | ||
| canonical_name | Accepted scientific name, including genus and/or species names | Pteropus alecto |
| common_names | Common or vernacular names for a species | Black flying-fox |
| kingdom | Taxonomic ranks (note: this is currently case-sensitive) | Animalia |
| phylum | Chordata | |
| class | Mammalia | |
| order | Chiroptera | |
| family | Pteropodidae | |
| genus | Pteropus | |
| regnum | Plantae | |
| division | Charophyta | |
| classis | Equisetopsida | |
| ordo | Fagales | |
| familia | Casuarinaceae | |
| Genomic data attributes | ||
| accession | Accession number assigned to a sequence datum by the source repository | GCA_900500725.1 |
| level | The assembly level for a genome: contig, scaffold, chromosome, complete genome | Scaffold |
| Specimen data attributes | ||
| institution_code | The code for the institution, such as a museum, where a specimen has been deposited. Most institution codes will follow the convention recorded in the Global Registry of Scientific Collections (GRSciColl) | AM |
| collection_code | The code for a specific collection within an institution | Ichthyology |
| recorded_by | The group or individual who originally collected the specimen | Sandy Desert Survey |
| identified_by | The group or individual who taxonomically identified the specimen | John Smith |
| collected_by | The group or individual who collected the specimen | John Smith |
| Other data attributes | ||
| data_source | Repository or database providing sequence or specimen data | NCBI Genbank |
| data_type | The type of data as defined in ARGA | Genome |
Complex searches can be built by combining search terms with standard query syntax. Examples of the most commonly used operators and expressions are provided below. A more comprehensive description of the syntax accepted by the Tantivy query parser used by ARGA can be found here.
- {John Smith} will return results where John Smith’s whole name remains intact as John Smith; searching for {J Smith} will broaden the results, returning results containing the terms {Smith} and {J}, e.g. J Smith, James Smith, Smith, J. and Katie Smith and John Silvs.
- {Macropus} will return any result containing the term Macropus, but {genus:Macropus} will return a result set of only species summary reports for species in the genus Macropus.
- {family:Macropodidae -Macropus} will return species summary reports for all macropod species, excluding any reports for species classified within Macropus (i.e. all non-Macropus macropod species).
- {Macropus AND institution_code:AM} will return only specimens from AM (Australian Museum) with Macropus in the scientific name.
Hint: Searches using specified attribute fields are case sensitive. All attribute terms should be written in lowercase, with words separated by underscores, and search value terms should be in proper case (e.g. Macropus, Genome).
Search results
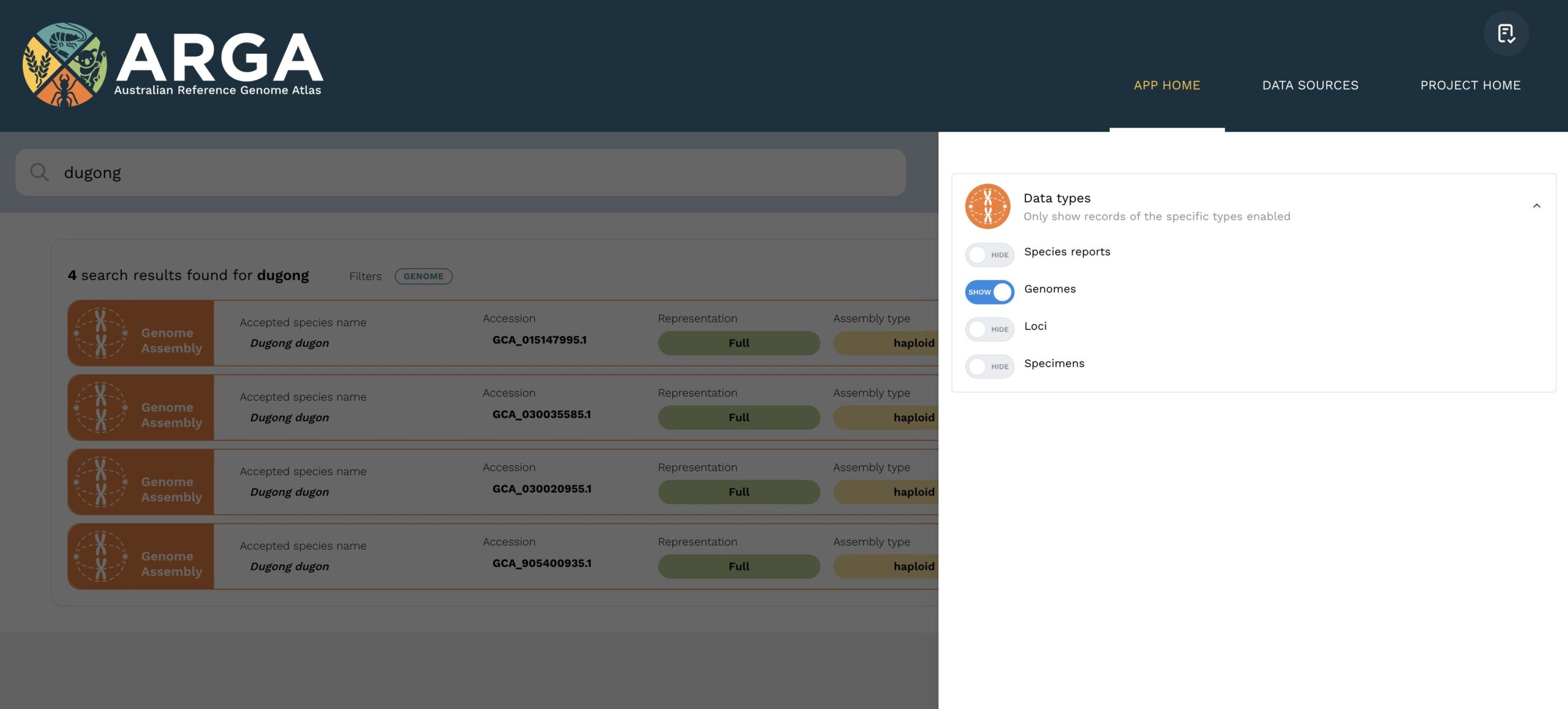
After pushing the {Enter} key (or {Return} key), the ARGA app will proceed to complete a search based on your input terms. The app will then direct you to a results page which lists the results of the search. Figure 1 shows a screenshot of the output in the results view page after searching for the term {dugong}. On any given results view page, there are four types of records which can be displayed: species reports, whole genome assemblies, single genetic loci, and specimens. You can use the filter icon at the top right of the results window to refine the displayed results according to a specific data type (see Figure 2).

The species report listing includes information about taxonomic classification and an overview of the number of genome assemblies and other genetic data records indexed for that species. Clicking the view button will take you to the complete species report, where you can explore the available data in more detail. The genome and locus listings include key metadata and the source repository from which the data were indexed. The view button opens the full data view page, where more metadata and provenance information are available. Specimen listings provide information about physical specimens which have been formally accessioned into registered collections and/or sampled to produce a genomic datum. You can go to the complete specimen view page to find out more about a specimen and any associated genomic data by clicking the view button.
Species reports
The primary aggregator used by ARGA to index genomics data, other genetic data and specimen data is the scientific species name. All species included in ARGA have a species report page, regardless of whether any genomic data are associated with that record. Species reports can be accessed from search results or browse pages. Different data types are presented in different tabs.
The header on the species report page includes the scientific name of the species, followed by icons indicating a higher taxonomic grouping (such as mammals or birds) and/or membership of a curated functional or ecological species group. You can hover over these icons for more information, or click on them to be directed to the higher taxon report for that grouping. At the right of the header block, you can see at a glance whether a reference genome is available for that species (i.e. whether it has a genome assembly on NCBI RefSeq), as indicated by a green tick or red cross.
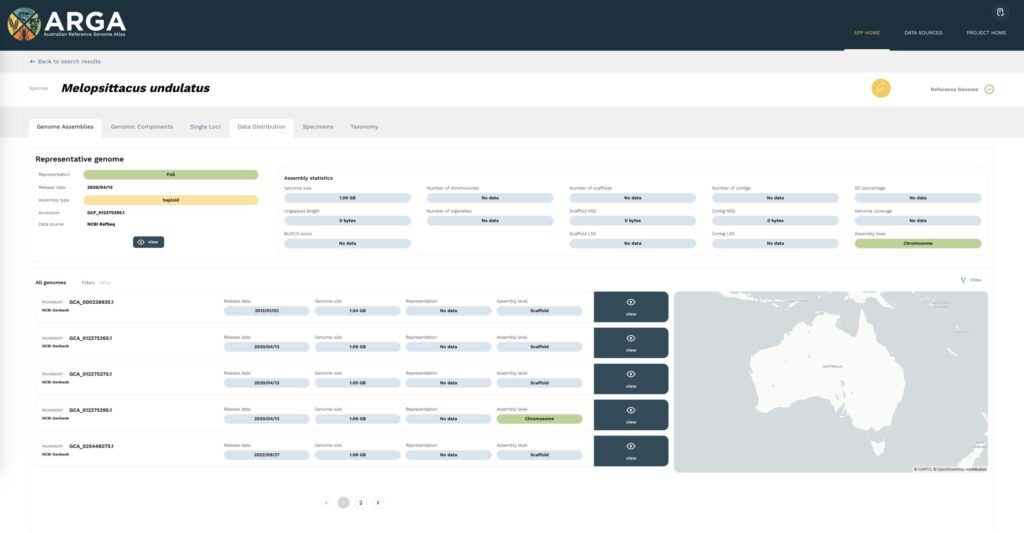
Genome assemblies
The genome assemblies tab (Figure 3) lists the nuclear genomes indexed for a given species. If a reference genome is available from NCBI RefSeq, this is designated at the top of the page as the representative genome, and the assembly statistics are provided. Other genome assemblies are listed below; these can be filtered according to assembly level, genome representation, and release type by using the filter icon at the top right. Where applicable, locations are plotted on the map according to the collection location of the specimen or sample from which the assembly data was generated. Clicking on the dark blue view icon beside a listing will bring you to the full data view page for that accession.
Genome assemblies: full data view
The full data view page for each of the indexed genomic assemblies (Figure 4) contains a selection of metadata indexed from the source repository. These are provided in the blocks titled genome details, assembly statistics and data provenance. The data availability block provides a rapid-glance overview of the types of metadata available for a datum within the ARGA index; green ticks indicate data presence and red crosses indicate data absence.

Within the original data box inside the genome details container, there are buttons to assist users to access original data. The first of these is the add to list button. This button saves a link to the datum for later access. Saved data can be viewed at any time by clicking on the white list icon at the top right of the global header (Figure 5). The number of items in the list is designated in a small orange circle beside the list icon (Figure 5). The links provided in the list will send you directly to the source repository to access the data. The get data button opens a new tab where you can download the sequence datum directly, whereas the go to source button directs you to the corresponding page for that accession in the source repository (where available). The send to Galaxy button is designed to send data directly to the online analysis platform, Galaxy Australia; this feature is in development and will be available in future ARGA implementations.


You can find more information about a genomic datum, including additional metadata relating to the collection of a specimen or sample, as well as its sampling, sequencing, and assembly, by clicking on the go to specimen button in the specimen information block. This button directs to the complete specimen view, which provides details about the events between the collection of a specimen and the deposition of a sequence datum and its reuse.
Genomic components
The genomic components tab (Figure 6) lists raw component data which are used by bioinformaticians in the assembly of whole genomes. The tab also lists other data types which are not captured on the genome assemblies and single loci tabs. Data found here may include raw reads, targeted enrichment sequence reads and reduced representation sequence reads (e.g. DArT data). The go to source button alongside each record directs to the relevant page at the source repository, where you can access more information about the data.

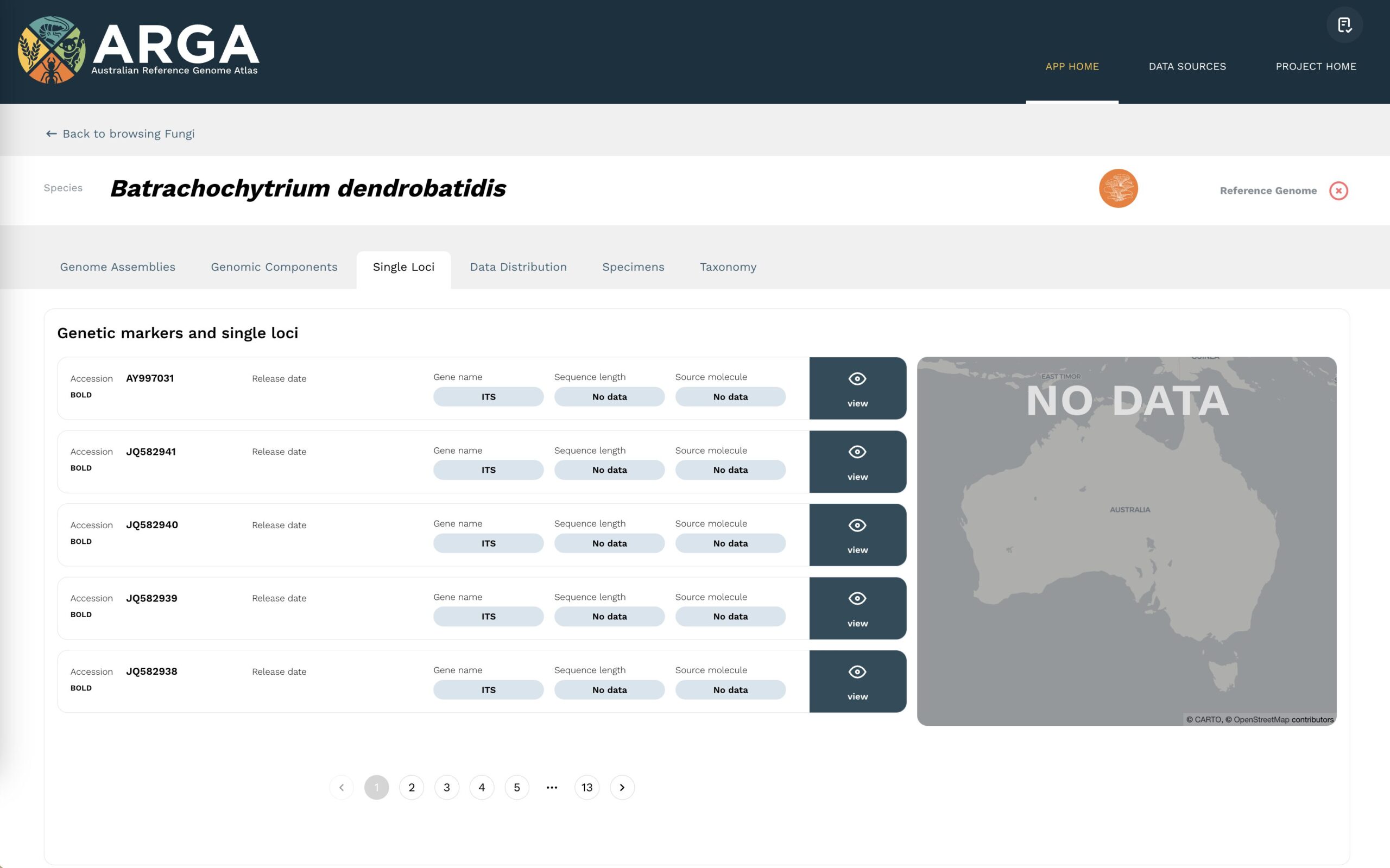
Single loci
Locus and marker sequences are listed under the single loci tab (Figure 7) for a species, along with the accession number (or alternative identifier), the locus (or gene) name, the sequence length, and the source molecule (e.g. DNA, RNA). Data are plotted on the map according to any geolocation co-ordinates provided for the collection of a specimen or sample. More details for each datum are available by clicking the corresponding ‘view’ button, which will take you to the single locus data view page.
Single genetic loci: full data view
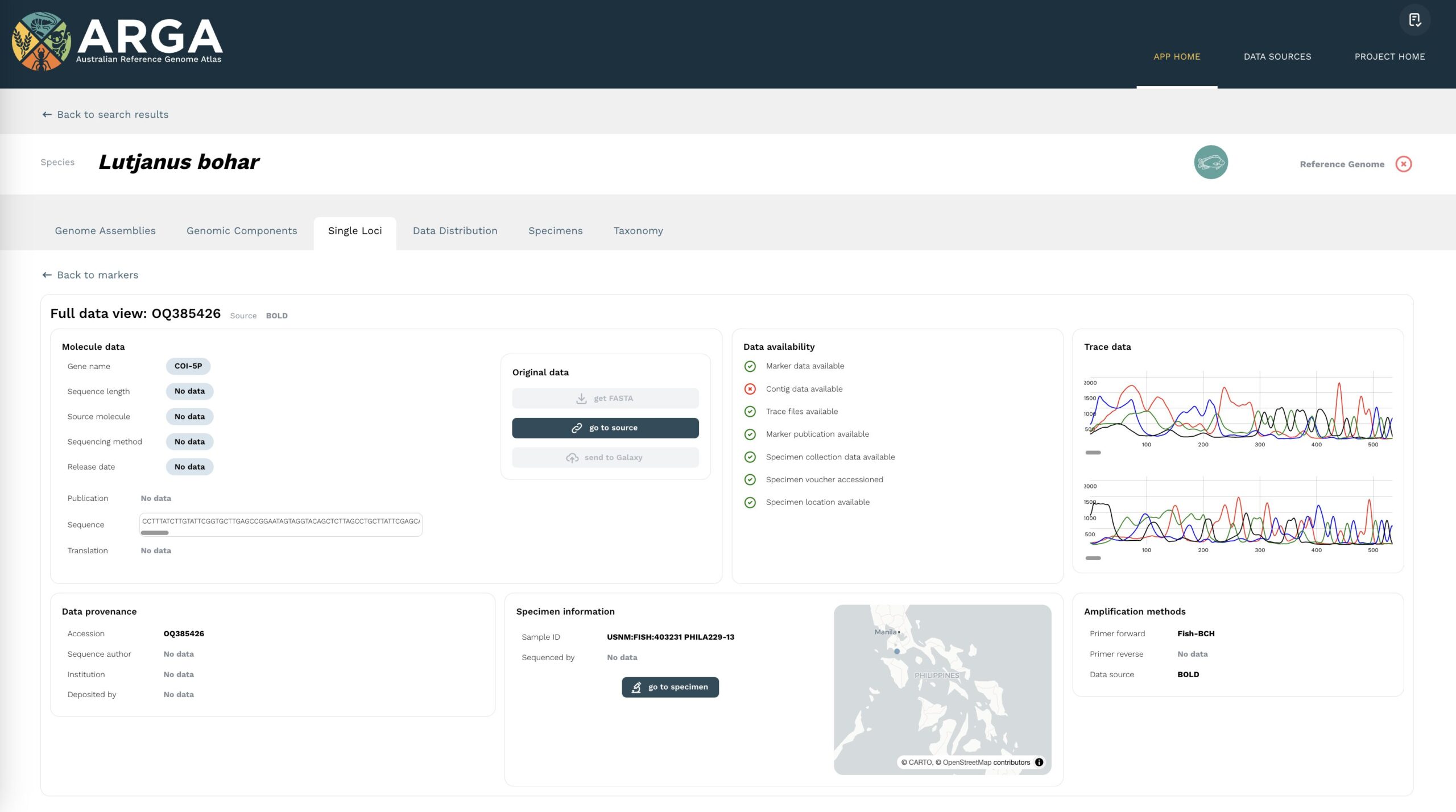
The full data view page for a single locus (Figure 8) contains a selection of metadata indexed from the source repository for single genetic locus data records. These metadata are organised into containers for molecule data, data provenance, and amplification methods. An overview of the total metadata available for a datum is provided in the data availability block, as indicated for rapid glancing by green ticks and red crosses.
Within the molecule data box, there are buttons to enable access to the raw data. The get FASTA button navigates to an external site for direct download of the sequence data. The go to source button directs to the corresponding page for that accession in the source repository. There is another button, send to Galaxy, to send data directly to the online analysis platform, Galaxy Australia; please note that this feature is in development and is not currently available. The page features a trace data viewer block which can be used to visualise the sequence electropherogram extracted from the relevant ABI file, if this is available. Where multiple files are attached to a single sequence, multiple electropherograms are shown and can be compared vertically.
You can find more information about a sequence datum, including additional metadata relating to the collection of a specimen or sample, as well as its sampling, sequencing, and assembly, by clicking on the go to specimen button in the specimen information block. This will take you to the complete specimen view page, with details describing the events between the collection of a specimen and the deposition of a sequence datum and its reuse.
Data distribution
The data distribution tab (Figure 9) features a map for visualising the geographic distribution of data indexed in ARGA for a given species. Points are plotted according to the coordinates given for the collection location of a specimen or field sample. You can click on any of these points to find a link to the relevant specimen or datum page, where further details are available. Some data have collection locations outside of Australia, and you may need to zoom in (or out) to navigate around the map to find them.
You can adjust the data types (whole genomes, loci, other data, and specimens) displayed on the map using the toggles on the right. The numbers beside each data type indicate the number of data points recorded with geo-coordinates over the total number of data points available for that data type within the ARGA index. Data without collection location geo co-ordinates are not displayed on the map.
Data are displayed as per the coordinates provided in the indexed metadata, however, it is important to note that coordinates may have been intentionally generalised (e.g. for sensitive species), or, in some instances, may have been incorrectly reported.
Overland regions highlighted in yellow and aquatic regions highlighted in blue designate the known range for that species in Australia, as sourced from the Australian Faunal Directory. These regions are defined according to the interim biogeographic regionalisation of Australia (IBRA) and integrated marine and coastal regionalisation of Australia (IMCRA), respectively. The relevant region names are listed under the Distribution heading.

Specimens
Records of physical specimens for a given species are listed under the specimens tab (Figure 10). Specimen data are sourced both from sequence data repositories and from records provided to the Atlas of Living Australia by registered collections, including museums and herbaria. Where programmatically feasible, data are aggregated according to specimen registration number, so that a specimen record pulls in any collection or accession information described in collection records as well as any sequencing information which has been attributed to that specimen. For genomic data indexed without a specimen registration number, a sample identifier (such as a biosample number) is used. For data without a sample identifier, a nominal registration number is generated by ARGA, unique to that datum.
In addition to the registration number, specimen records are listed alongside collection location information, type status (designating whether a specimen is considered a holotype, paratype, or other form of type), and whether derived genomic data are available. For records with geo co-ordinate data, specimen collection locations are shown on the map. Clicking the view button will direct you to the complete specimen view for that record.

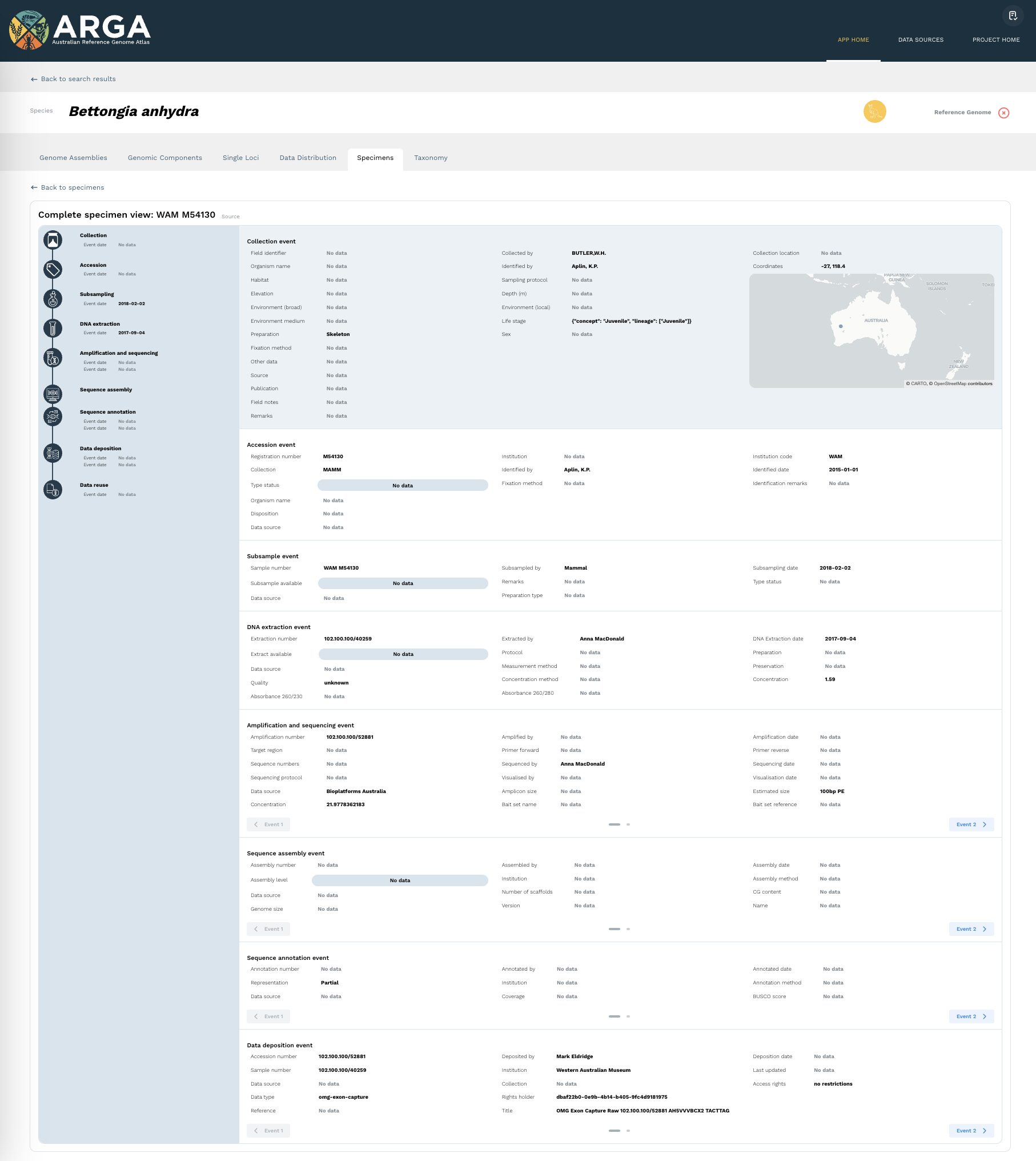
Complete specimen view
The complete specimen view page (Figure 11) presents information relating to each event between the collection of a specimen and deposition of a genomic datum and its reuse. Data on this page may have been sourced from metadata attached to indexed genomic data and/or from records supplied by formal collections. The data source is provided separately for each event. The fields listed under each event have been chosen to accommodate a wide range of information and detailed processes. However, sequence data are rarely deposited with a full complement of relevant metadata. As a result, many of the specimens listed in ARGA will have little data available, and some event stages will have no data at all.
Sometimes, multiple locus sequences and/or assemblies will have been derived from a single specimen. In these cases, there may be more than one event recorded at each event stage (e.g. amplification and sequencing event). You can navigate between these different events by clicking on the arrows at the bottom left and bottom right of that event block.
Taxonomy
A full taxonomic classification of a species is provided on the taxonomy tab (Figure 12). This view includes the scientific name, the taxonomic status of that name, and common names. There is a higher classification block providing details of the placement of the species within each level of a taxonomic hierarchy; the source for the classification shown is indicated. Clicking on the buttons for any of the higher taxa directs to the higher taxon summary and browse page for that specific higher taxon.
It should be noted that different repositories and databases may employ different classification structures (in terms of both ranks and attributions of groups within those ranks), and that individual genomic data may not always be updated to account for taxonomic revisions or re-identifications. ARGA indexes data according to the taxonomic names defined in the source repository. These names are matched to an ARGA names index, which is derived largely from the taxonomy as defined in the Australian National Species List and Australian Faunal Directory, and as reflected in the taxonomy of the Atlas of Living Australia; the ARGA names index is supplemented with additional species external to Australia (such as those with biosecurity implications) and other species-like taxonomic units (like molecular operational taxonomic units (= MOTUs)). For each taxon, the source used to assign taxonomy is provided on the taxonomy tab, and can be accessed by clicking on the hyperlink.
Browsing
Users can scroll through the data indexed on ARGA by clicking on the browse icons on the homepage. Browsing categories are divided into data types, taxonomic groups, and functional or ecological groups. Clicking on any one of the icons on the app home page launches the browse page for that chosen group.
The browse page feature creates a card for every species, including an image (if available), the scientific name, and an indication of whether one or more genome or locus sequences have been indexed for that species. The presence of data for the species is designated by a green tick, and the absence of data in the index is shown by a red cross. Clicking on a card opens the species report page. Users can return to browsing the set of cards by using the back arrow, which is located on the species report above the taxonomic name.
On any browse page, the selection of species displayed can be adjusted by clicking on the filter icon at the top right, and turning on and off various filter combinations which are provided under the filter submenus. Please note that applying the filters does not adjust the statistics in the data summary provided on the top half of the page (currently implemented for taxonomic groupings only).
Browsing by data type
The first set of icons on the app home page enables browsing by the type of data contained in the index. Users can browse sets of species for which there are genome assemblies, single genetic loci and registered specimen records (see Figure 13). The number of data records is calculated below each particular data type. Each set of species can be browsed by clicking on the selected icon.

Browsing by taxonomic groups (and higher taxon reports)
The ARGA interface presents the opportunity for browsing species by higher-level taxonomic groups (e.g. plants, animals, or fungi) to see overviews of the data in the index, and for scrolling through the species contained within that higher taxon. Higher taxon reports (Figure 14) can be accessed from the ARGA app home page by clicking on the icon relevant to the group of interest. They can also be reached by clicking on a taxon of interest from within the taxonomy tab of any species report.
Hint: You can also jump to data summary pages directly by typing in the URL using the formulation: {{app.arga.org.au/} + {rank/} + {Taxon}} (making sure to capitalise the taxon name). For example, to go straight to the summary page for beetles (Order Coleoptera), type in {app.arga.org.au/order/Coleoptera}.
The data summary block provides graphs and statistics describing the availability of indexed genomic data for the species within a designated taxonomic rank. The total number of species included in the taxon is given in the taxonomic breakdown block to the right. These species are listed on the browse cards below. Rank-specific data summaries are available for all eukaryotic taxa at the major Linnaean ranks. From the data summary page for a given taxon, you can navigate to summaries for higher ranks in the classification hierarchy by clicking on the scientific names in the higher classification block above the graphs.
Browsing by functional and ecological groups
The functional and ecological groups each contain lists of species curated by ARGA to reflect a particular theme (e.g. poisonous and venomous species, or species vulnerable to bushfires). These species lists have been built from authoritative sources and are fully referenced on the Data Sources page.
Data sources and citations
ARGA indexes and uses Data from multiple online sources, which are listed on the Data Sources page. Users can navigate to this page by clicking on the tab marked Data sources on the global header at the top of the ARGA app. Data sources are provided with hyperlinks and, where relevant, citations. If users reuse any data found on ARGA in any published work (online or in print), they must reference the primary source work, including the author and/or data provider with literature citations (where applicable). Please refer to the ARGA Data Use Policy for more information.
Citing ARGA
Please cite ARGA by including the following statement when publishing your research:

This research was supported by the use of the Australian Reference Genome Atlas (ARGA), an NCRIS-enabled platform powered by the Atlas of Living Australia (ALA), in collaboration with Bioplatforms Australia and the Australian BioCommons, and supported by the Australian Research Data Commons (ARDC).
Contact us
For technical assistance with using the ARGA app, please send any questions to . We welcome any feedback or issues and bugs reporting via this address also.
For other general enquiries about the ARGA Project, please direct any questions or feedback to .
Page attributions
Quill With Ink icon by Icons8